Psychology: Physiological psychology: Taxi drivers brain activity
Authors: Maguire et al. (1997)
Key term: Taxi drivers
Background/ Context: The invention of technology such as PET and MRI scanners has led to many studies on brain regions and activity. Topographical memory is the knowledge of landscapes and the spatial relationship between them - a cognitive (mental) map we have inside our heads. Maguire's study is on semantic topographical memory and what brain regions are responsible for it.
PET: Positron Emission Tomography.
Semantic memory: General world knowledge we have
Aim/ Hypothesis;
- Confirm findings of other studies that specific brain regions are responsible for semantic topographical memory retrieval.
- Determine which specific region of the brain is responsible for landmark knowledge when location information is not needed (topographical memory without sequencing).
- Study topographical and non-topographical semantic memory to see if there is brain region that is activated for both.
Method: Laboratory experiment with pre-study questionnaire
- Participants filled out a questionnaire asking them about:
-Areas of London they were familiar with.
-Films they were familiar with from a list of 150 films from 1939 to the present day.
-20 world famous landmarks participants could visualize but had never visited.
- This would let experimenters choose the most appropriate routes, landmarks and films they wanted the participants to describe.
- Pilot study done with non-participating taxi drivers before this experiment to make sure the tasks and equipment were fit for the study.
Variables:
Independent variables
- Main variables: Topographical and Sequencing.
- The participants completed 6 tasks, each done twice. However only 5 of those tasks are used in this study.
- Baseline task - Repeat two sets of 4 digit numbers (to compare simple level of activity to the 4 other tasks).
- Describe the shortest route from one point to another in the City of London (same route for all participants, one they were familiar with) (topographical and sequencing).
- Describe a landmark in terms of features, appearance, etc (Same landmark for all participants, one they all knew) (topographical and non-sequencing).
- Describe the plot of a famous film between two given points of the film. (familiar to all participants) (non-topographical and sequencing).
- Describe individual frames of some famous films eg. imagery, characters, etc. (familiar to all participants) (non-topographical and non-sequencing).
Dependent variable: Region/s of brain
activated.

Design: Repeated measures -
Factorial design
- Orders of tasks randomized for each participant and counterbalanced against each other.
Participants and sampling technique:
- 11 right handed male taxi drivers.
- Minimum of 3 years experience driving, average was 12.5 years.
- Average age 45 years.
Apparatus:
- London route from Grosvenor Square to the elephant and castle.
- World famous landmarks
- Films with still frames
- Siemens PET scanner
- H2 15 O bolus (radioactive isotope for scanning) and saline flush to remove isotope from body.
Controls:
- Landmarks chosen were places participants never visited in person, so they were not sequential memories.
- Films chosen had been watched by participants at least 5 times, so they were all familiar with them.
- Scanner procedure and injections given were the same for each participant.
- Pilot study done to ensure all tasks are fit for purpose.
Procedure:
- Participant filled out pre-study questionnaire.
- Participant arrived at the laboratory and cannula (tube for delivering fluid) was inserted in his arm. Participant was blindfolded and placed in scanner.
- Participant received H2 15O bolus injection through cannula (which takes 20 seconds to enter bloodstream). This was followed by a saline flush (which also takes 20 seconds).
- Scan frame time was 90 seconds (H2 15O has a half life of 2 minutes). 1 of the 5 tasks was presented and the participant had to give a description of the route, landmark, film plot or frame, which was digitally recorded. MRI was also taken for each taxi driver,
- After the task was completed their would be an 8 minute wait before another task was done.
- Steps 3, 4 and 5 repeated 12 times (2 times for each study including another task for another study not in this paper).
- Participant was debriefed and study was complete.
Data: Quantitative data - rCBF (Regional cerebral blood flow) would flow to areas of brain activated, which would be recorded by the PET scanner, interpreted and analysed. 3D images of the PET scanner show exactly which brain region is involved in each process.
Findings:
- No difference in amount of speech per participant (around 50 seconds).
- Very few differences in route chosen by participants.
- Routes (T+ S+): Right hippocampus, parahippocampal gyrus, cerebellum, media parietal lobe, posterior cingulate cortex, extrastriate regions.
- Landmarks (T+ S-): Parahippocampal gyrus, posterior cingulated cortex, medial parietal lobe, occipito-temporal region, cerebellum.
- Films (plot and frames - T- S+ and T- S-): Left frontal region, cerebellum, middle temporal gyrus, left angular gyrus.
- Cerebellum activated in all activities.
- The parahippocampal gyrus and medial parietal lobe were used for both topographical activities. However, the right hippocampus was only activated in the route activity - topographical and sequencing.
Conclusion:
- Regions of topographical semantic memory similar as those in previous studies.
- Right hippocampus used for sequential route planning.
Strengths:
- Laboratory experiment - High level of control - pilot questionnaire, routes, landmarks and films chosen - easily replicated to test for reliability.
- Laboratory experiment - High level of control - confidence the type of tasks directly affected brain function - IV directly affects DV - baseline task, type of tasks.
- Technological equipment - lack of human error
- Testing physiological aspects (brain function) - can be generalized to wider population, as majority of humans have similar brain structures.
Weaknesses:
- Laboratory experiment - Artificial setting - Participants were tested whilst undergoing a PET scan, which is not a normal setting for recalling memory, therefore this experiment may not have ecological validity
- Laboratory experiment - Lacks mundane realism - Participants were blindfolded and asked to recall memories during a PET scan, which is not a normal task people do in daily life.
- Can't be generalized - Participants were all taxi drivers - It is possible the right hippocampus in sequential topographical memories is only crucial in taxi drivers.


 Simple diagram of transcription.
Simple diagram of transcription. Ribosome.
Ribosome. tRNA molecule.
tRNA molecule. Ribosome in translation.
Ribosome in translation. Translation diagram.
Translation diagram.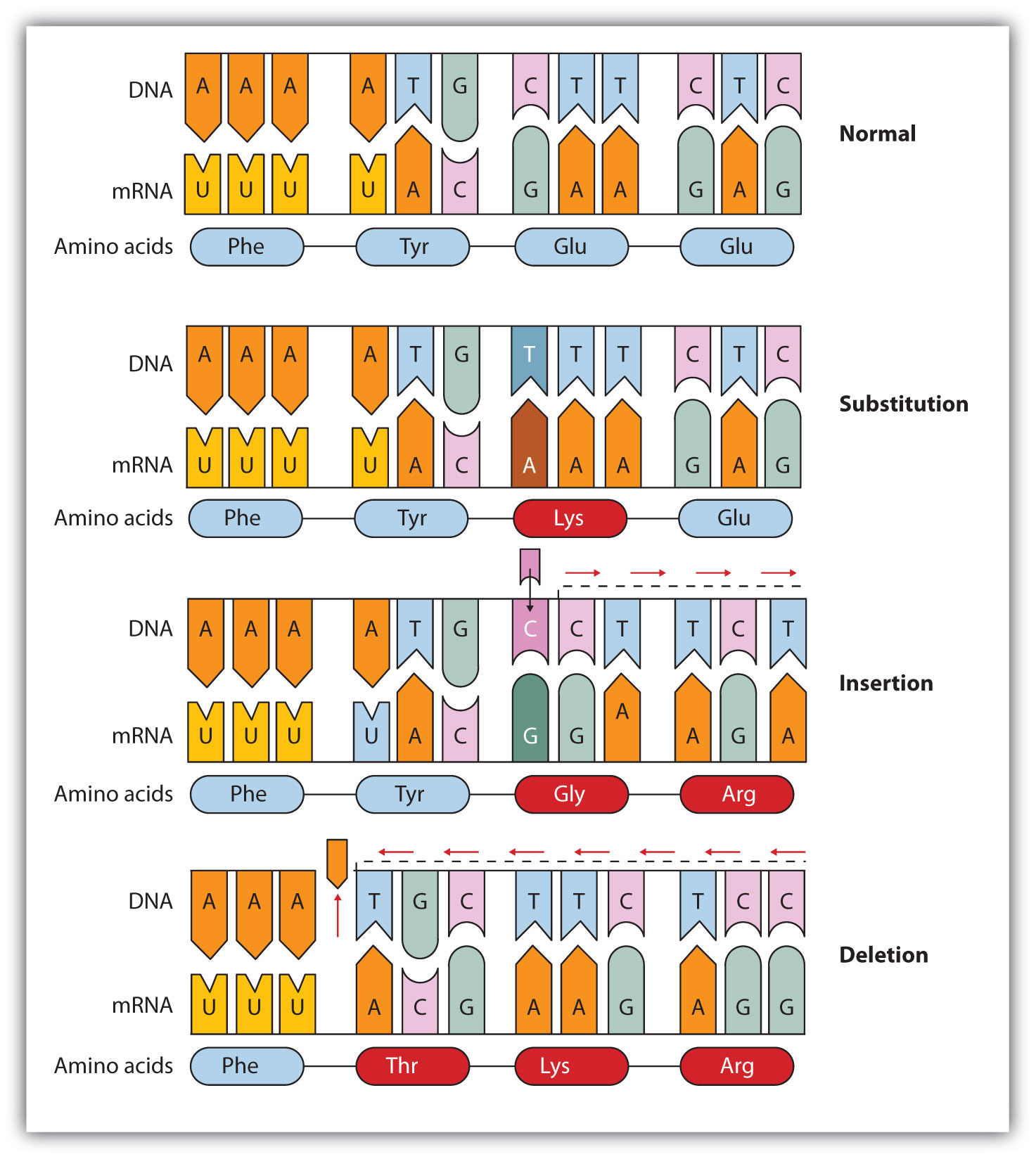 DNA mutations.
DNA mutations. Simple diagram of semi-conservative replication in DNA.
Simple diagram of semi-conservative replication in DNA.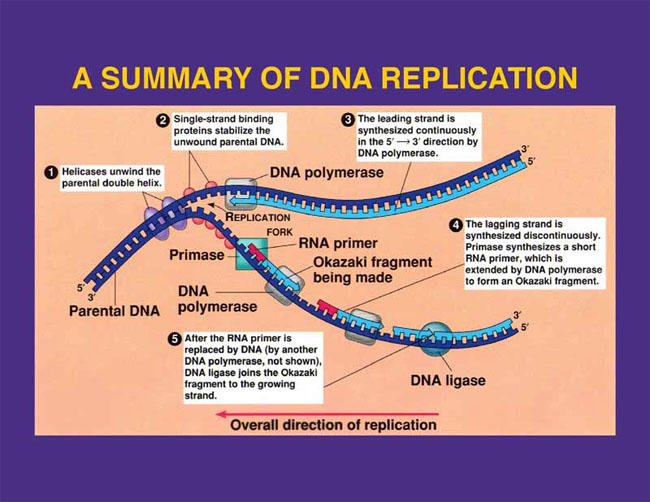
 Structure of nucleotide.
Structure of nucleotide. Structural formulae of the nitrogenous bases.
Structural formulae of the nitrogenous bases.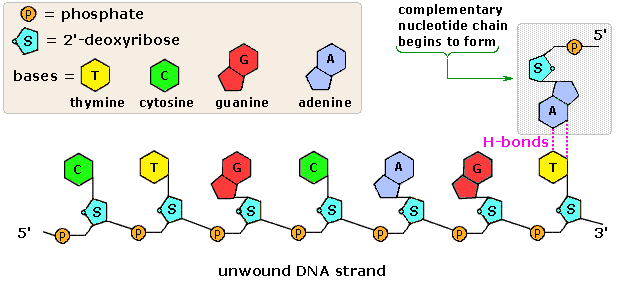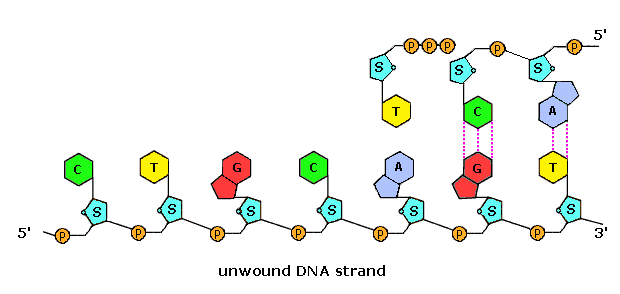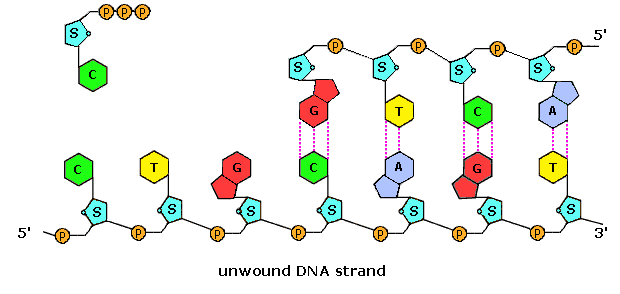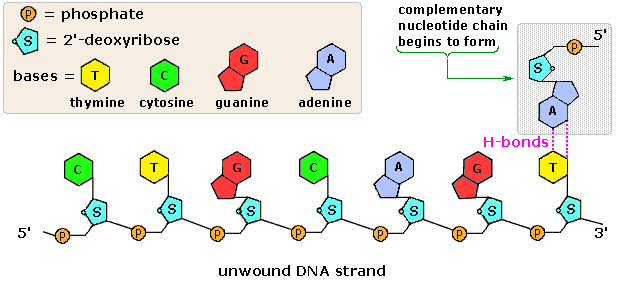 Animation of DNA replication and the DNA strand.
Animation of DNA replication and the DNA strand.